I needed the location of nucleosomal dyads in the K562 cell line (ENCODE tier 1 line). Surprisingly, although plenty of MNase-seq data for that cell line is available, no nucleosome and dyad prediction exists.
NuMap
I found the NuMap software by Anton Valouev to do exactly what I intended.
Since it is in a somewhat obscure page and this seemed to be the only place where this software was, I have uploaded it into a Github repository for the sake of preservation.
Predicting dyads from MNase-seq data with NuMap seemed trivial: I downloaded the K562 MNase-seq data set (11 replicates ~85Gb!!), combined all replicates and ran NuMap on the data(instructions on the Github README).
From NuMap output there are dyad positions in bed format and you can also produce several metrics to evaluate how good the prediction was.
Distograms & phasograms
Valouev describes two measurements of the frequencies of distances between MNase-seq reads. The frequency of distances between reads mapping to opposite strands can be used to build a “distogram”, which ilustrates the expected nucleosome length (147 bp) - this is consistent across most eukaryotic cells. The frequency of distances between reads mapping to the same strand gives a measurement of the distance between nucleosomes, as they’re separated by some linker DNA - (Valouev calls this plot a “phasogram”). This measurement, on the other hand tends to be species and cell-type specific.
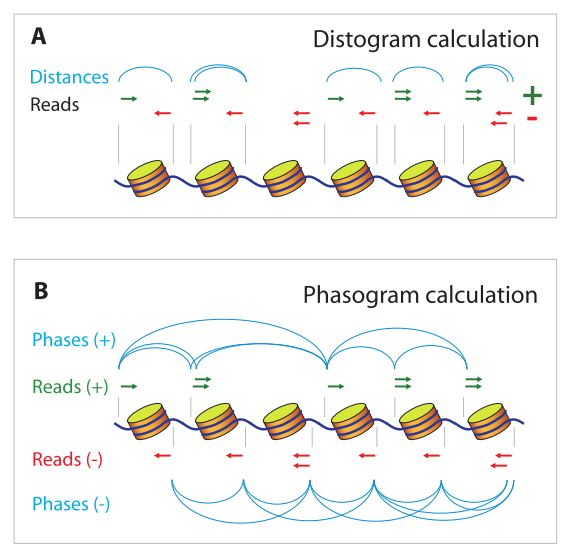
Valouev, A., Johnson, S. M., Boyd, S. D., Smith, C. L., Fire, A. Z., Sidow, A. (2011). Determinants of nucleosome organization in primary human cells. Nature, 474(7352), 516–520. https://doi.org/10.1038/nature10002
K562 predictions:
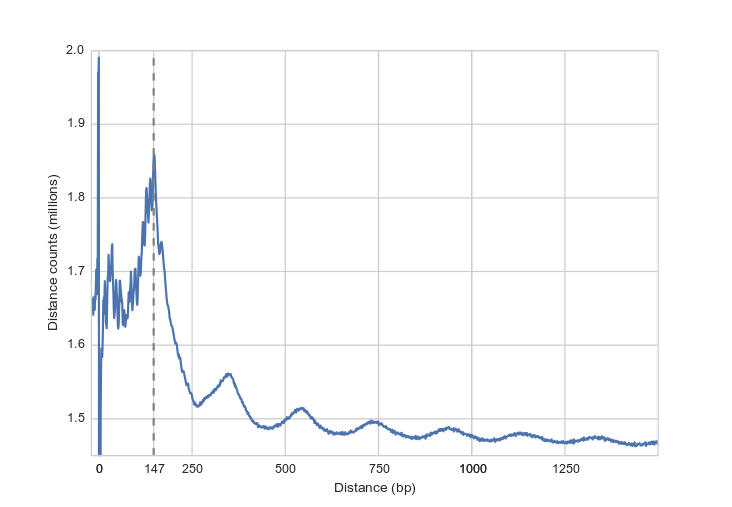
The expected 147 bp nucleosome length in K562 cells.
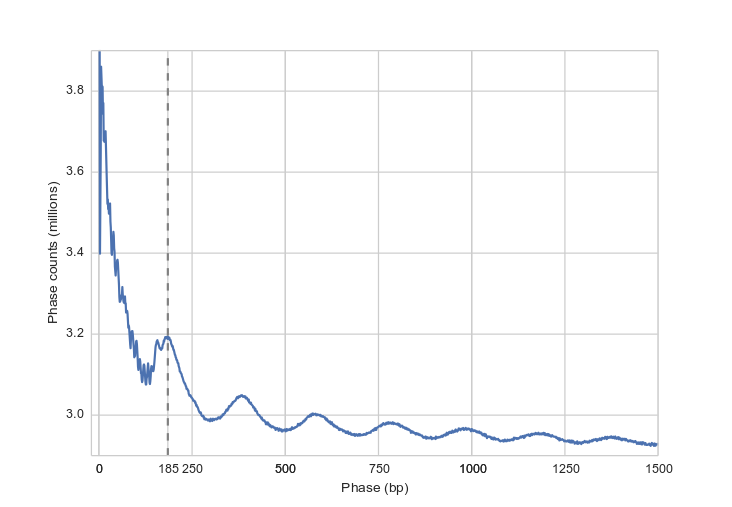
The average distance between dyads in K562 cells seems to be 185 bp.